Today I’ll try to summarize another paper that is difficult for one reason: figuring out how the authors (including Nobel laureate Svante Pääbo) sussed out which genes originated in Neanderthals and Denisovans (both lineages now extinct) and which in their sister lineage: the separate lineage leading to modern humans. (All three lineages shared a common ancestor.) As we know, the modern human genome contains a lower percentage of genes that originated in Neanderthals, and this is also true for genes that originated in Denisovans (the latter are found more often in modern populations from Oceania, Asia, and in native North Americans).
As for the two extinct lineages, both derived from a single ancestor that, according to the paper below, left Africa for Eurasia about 600,000 years ago. That traveling lineage then split at an uncertain time to give rise to the Neanderthals, who went extinct about 35,000 years ago, and to the Denisovans (known from but a handful of teeth and bones), who went extinct around the same time as did Neanderthals. In the meantime, the lineage that gave rise to us—”modern” humans—stayed in East Africa, leaving for Eurasia about 60,000 years ago, and some individuals in this group lived near the already-present Neanderthals and Denisovans.
Although human paleobiologists, who love to identify new species, call the Denisovans and Neanderthals species different from modern humans (i.e, different from “Homo sapiens“), I’m stubborn and consider all three groups members of the same biological species. That’s because there’s evidence of gene flow among all the groups: from Neanderthals and Denisovans to modern humans, from modern humans to Neanderthals, and even from Denisovans to Neanderthals and vice versa. Because these archaic genes persist in modern humans, the hybrids between the lineages must have been fertile to allow such backcrossing. Since we have populations who lived at least partly in the same area and produced fertile hybrids, they can be considered biological species, though perhaps biological species in statu nascendi.
Here’s a diagram of the three lineages from the paper whose title is below. The arrows show the direction of gene exchange and some examples of variants transferred by hybridization.
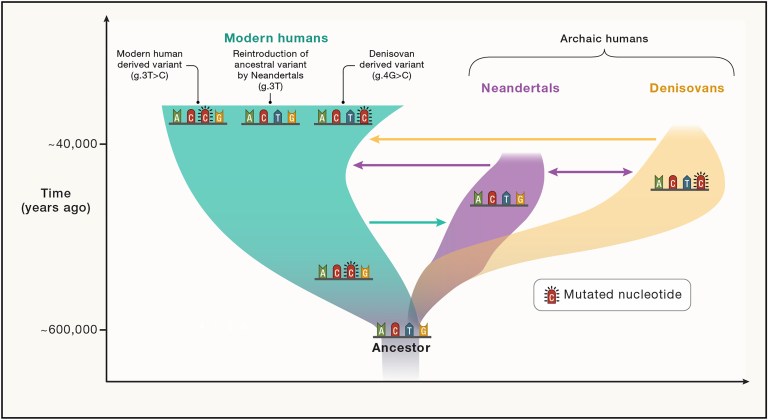
You can read the paper below by clicking on the title. The object of the paper is to answer this question:
Which genes were transferred between the lineages, and what effect did they have on the individuals who carried them?
Now, of course, the first question is “How do we know which genes actually originated in one of the three lineages after it split off from the others, and how do we know in which direction that gene was transferred to another lineage.” This is not an easy question, and I asked my friend Phil Ward, an entomologist and systematist who works at UC Davis. I’ve put his explanation below the fold in case you want to know.
I trust the authors’ determinations of which genes originated where, and which ones were introduced into a given lineage by hybridization; after all, this is Pääbo and his group! So I’ll just give a list of a few of genes transferred, which way they went, and what they appear to do in modern populations. Let me add two things. First, while it’s easy to find out what a gene does (it can be tested in cell culture or by inserting it in mice), it’s not so easy to determine what effect it has on the phenotype or reproduction of modern humans.
Second, the paper is quite “adaptationist”, with the authors suggesting reasons why a transfer might have been adaptive. (If it was really bad for the carrier, it would have disappeared from the population.) However, very few of the transferred genes are present in very high frequency in modern humans or Neanderthals, and so, if they had a really beneficial effect on reproduction or survival, one would expect that they would be “fixed” (present in every individual) or at least high frequency. Since that’s not usually the case, the authors float hypotheses that transferred genes are good in some populations but not others. This seems to some extent like post facto rationalization based on a diehard adaptive viewpoint.
On to the genes! Indented sections are mine except for doubly-indented sections, which are excerpts from the paper:
Mutations in the Neanderthal lineage that got in modern humans via hybridization.
Genes affecting fatty acid and lipid metabolism. These genes appear to increase the risk of type 2 diabetes, so they’re not good for us.
Genes increasing sensitivity to pain (a sodium-channel gene in the nervous system). This gene appears to have a salubrious effect, because it’s associated with an increase in lifespan (genes that decrease pain sensitivity can allow you to suffer injuries and infections without noticing them, which is why sufferers from Hansen’s disease, lacking pain sensitivity, lose digits and other body parts). But if the gene is so good for us, why is it present in so few of us (0.4% of people in the UK)?
Genes affecting gestation. We received a progesterone receptor-variant that mutated in Neanderthals and that actually increases the frequency of premature births. Here’s how the authors explain its persistence at high frequencies in both Neanderthals and modern human populations (some of the latter have a frequency of the gene of 20% or higher):
Since [this variant] is associated with an increased risk of premature births in present-day humans, it has been suggested to represent an evolutionary disadvantage to Neandertals, especially in the absence of modern medical care. However, the Neandertal variants are also associated with an approximately 15% decreased risk for bleeding and miscarriages early in pregnancy as well as with having more siblings. It is therefore tempting to speculate that it represents an evolutionary trade-off where the Neandertal variants rescues pregnancies that would otherwise have resulted in miscarriages, but the price paid is that some of these pregnancies result in premature births. Notably, two different versions of the Neandertal progesterone receptor gene have been contributed to modern humans, and both have risen in frequency, as shown by an increase in their occurrence in skeletal remains of individuals over the past 10,000 years. Both Neandertal versions result in higher expression of the progesterone receptor and may thus mediate a higher progesterone effect during pregnancies. This is compatible with the finding that progesterone administration lowers miscarriage rates in women who previously experienced miscarriages and suggests that increased progesterone effects mediated either by higher hormone levels or by higher receptor levels may protect at-risk pregnancies.
It’s okay to speculate, but perhaps there are other effects of the gene that we don’t know about, and are the bad effects of premature births overcome by the beneficial effects on bleeding and reduced early miscarriage? What we have here is a reflection of the author’s view that the transferred genes must in general have a net positive effect on reproduction, even if they can’t demonstrate it.
Genes affecting the immune system. Many of the genes we got from Neanderthals appear to interact with viruses and are at high frequencies in humans; the authors thus speculate that they spread to ward off infections and still do so in modern populations. Also, some of the variants have big differences in frequency between populations, which the authors attribute to population-specific infections. Further, some of the variants may cause autoimmune disease (again, we know little about their effects on modern humans, which may be small.) But they speculate that the existence of so many Neanderthal variants in modern humans strongly suggest that they spread in our lineage via selection for disease resistance.
Mutations in the Denisovan lineage that got in modern humans via hybridization.
Genes affecting adaptation to high altitude. Here, taken from the paper, is the best example of a gene entering the modern human genome that is likely to have spread by natural selection. This example is pretty well known.
High altitude adaptation
One striking example of Denisovan influence on present-day populations is a 33-kb Denisovan DNA segment on chromosome 2 that occurs at an allele frequency of over 80% among Tibetans, while being absent or very rare in other Asian populations. It encodes EPAS1, a transcription factor induced by hypoxia that is involved in adaptation to low oxygen levels. Denisovans were present on the Tibetan high plateau; some of them may thus have been adapted to life at high altitudes and presumably contributed this genetic predisposition to modern humans as they arrived in the region.
We also received genes involved in producing adaptation to low temperature by “inducing brown fat”: these too seem to have spread in cold-climate populations by natural selection:
Cold adaptation and facial morphology:
Another example of a Denisovan genetic contribution is a 28-kb segment on chromosome 1, carrying the genes WARS and TBX15. It is present in almost 100% of Greenlandic Inuit and several other populations. The Denisovan variants affect the expression of genes that may influence adaptation to low temperatures, possibly by inducing brown fat.
At the end, the authors discuss genes that emerged in modern humans and found their way into Neanderthal lineages, including variants affecting purine (nucleotide) biosynthesis, preventing oxidative stress, gene splicing, and chromosome segregation. The authors then present a “combinatorial view” of the modern human genome, noting that we’re likely to contain a variety of variants coming from our now-extinct ancestors, but different modern individuals have different combinations. Here’s that view from the paper’s abstract:
We propose that the genetic basis of what constitutes a modern human is best thought of as a combination of genetic features, where perhaps none of them is present in each and every present-day individual.
_________
Reference: Zeberg H, Jakobsson M, Pääbo S. The genetic changes that shaped Neandertals, Denisovans, and modern humans. Cell. 2024 Feb 14:S0092-8674(23)01403-4. doi: 10.1016/j.cell.2023.12.029. Epub ahead of print. PMID: 38367615.
Click “read more” to see the method for determining where a mutation originated and which way it was transferred:
Explanation by Phil Ward:
Perhaps the most reader-friendly explanation is to be found in David Reich’s 2018 book “Who we are and how we got here. Ancient DNA and the new science of the human past” (New York, Penguin Random House). On pages 34 et seq. of this book, Reich introduces the “Four Population Test” (his term for the ABBA-BABA test) in which he notes that if we consider derived alleles present in Neanderthals, these are shared more with Eurasian populations than with African populations, consistent with Neanderthals interbreeding with the ancestors of today’s Eurasians—after migration out of Africa. In most cases when you have a derived Neanderthal allele (i.e., one not shared with the chimpanzee) then it is not present in any human population, but if it is shared with one of two human samples, that sample is more likely to be Eurasian than African.
How about the direction of gene flow? Green et al. (2010: 218) note that Eurasian humans are more closely similar genetically to Yoruba than to San, to a degree than can be measured by the D-statistic. As they put it, “D(P, San, Q, chimpanzee) is 1.47 to 1.68 times greater than D(P, Yoruba, Q, chimpanzee), where P and Q are non-Africans”. If the direction of gene flow had been Eurasian to Neanderthal, then there should be a comparable difference in the D statistics comparing Neanderthal to San and Yoruba. Again quoting Green et al: “D(P, San, Neandertal, chimpanzee) should be greater than D(P, Yoruba, Neandertal, chimpanzee) by the same amount”. But this is not the case: the ratio of those two D statistics is approximately 1.0.
Putting this more simply (and perhaps oversimplifying), if Neanderthals had acquired a significant amount of DNA from Eurasians, this should be reflected in a statistically greater genetic distance of their DNA to San than to Yoruba, just as Eurasian DNA shows. Since that is not the case, one can conclude that most of the Neanderthal-Eurasian gene flow was in the opposite direction: Neanderthal to Eurasian.


Very interesting. Great to read with my morning coffee! Thanks Drs. PCC(e) and Ward for this. Why I come to WEIT.
You are on a role today! I can barely keep up with the science posts here. 🙂
Well, he’s in a role and on a roll! 😉
There’s a lobster joke in there somewhere…
A man complains to the waiter.
“Why is this called oasis soup? It seems like it’s just ordinary tomato.”
The waiter says (or sings) “well, you got a roll with it.”
Frog dissection below:
Just checking if anyone pays attention 🙂
It is impressive enough to be able to sequence genomes from such ancient fragments of teeth and bone, but to then be able to carry out these genetic analyses, and draw such profound conclusions, is truly awesome and uplifting. What a nice contrast to the rest of today’s news.
Very interesting stuff. It’s nice for PCC(E) to point where sometimes the authors may be getting carried away with their speculations (just a bit, anyway), which helps to decrease our own risk for getting carried away with overly adaptationist assumptions. This is an invaluable service, since few of us have the expertise to judge such things very well for ourselves. So, thank you PCC(E)
+1
This was great. Thanks for the clear explanations. So cool!
This is fascinating. I’ve been reading a lot about genetics and the discoveries of the past two decades. Amazing stuff that changes so much with each passing year.
Very interesting summary from our dear host, and his take that Neanderthals, Denisovans, and us should all be considered part of the same species made me think. It would be incredible if these populations (Neanderthals and Denisovans) still existed!
I haven’t done 23and me or any other gene analyses on myself, and I’m not really interested, except for their “Ancestry package” that looks for Neanderthal (but not Denisovan) genes. I don’t know how much it costs, but I would be interested. After reading this post, I’m even more interested…enough to actually order a test? Probably not.
Either way, thanks for this excellent and most interesting post. And thanks for all the science posts of late!
I think I sort of understand this–thanks PCCE!
Exactly my response as well.
Fun stuff – thanks.
I highly recommend John Hawks website (johnhawks.net)
for on-going info on paleo-anthropology.
Two great science posts in one day! The readers of this website are truly blessed! I particularly appreciate our host’s critical but constructive comments on this paper. A lot of site owners would be content with simply reproducing it without contributing anything themselves.
It’s been about 40 years since I took Human Evolution. It’s amazing the things we can do now besides look at bones.
Very interesting. Thank you for your expert input.
Fascinating. I’ve been following this “story” since the earliest work by Svante Pääbo and his team yielded results that first suggested interbreeding between modern humans and Neanderthal.
Something I found very interesting, still do, is how much of the Neanderthal genome is still around. I had assumed there were just a relative few genes, but a number of studies looking at this question suggest, if I’m remembering the numbers correctly, between 40% to 70% (a range of results from different studies) of the Neanderthal genome still exists, scattered in small bits among modern humans. I’ve not come across such estimates for the Denisovan genome, probably because there is considerably less data about the Denisovan genome to work with. But maybe that’s changed in more recent years?
By the way, minor mistake in the title to the Denisovan section, “Mutations in the Neanderthal lineage” was accidently repeated, should be Denisovan.
I thought I was genetically Jewish because everyone told me that I both look and act like a Jew, so I took a 23 and Me test. It was revelatory. Not only am I a pure blooded Honkey, but I also have almost twice the percentage of Neanderthal genes of the average person. I look in the mirror and there he is. I am fuzzy headed, big snooted, short, and powerfully built with a deep chest and strong arms. Despite my small stature, my cranial capacity is in the top 98%. I have an obsessional urge to wander, never get lost, and am driven by a keen hunting instinct which explains why I am a naturalist. It is good to learn that despite my hybrid heritage I can still be considered human!
IIRC, some studies conclude that Neanderthal skull capacity was (or perhaps *is*) greater than that of modern humans. It must be nice to be in the top 98%. (Actually my guess is that you meant to say the top 2%, no?)
I actually wondered how (or why?) someone would know their skull capacity? WTF? Liked Bruce’s post, just sayin’…
The rehabilitation of phrenology? With modern imaging you could assess the lumps and bumps on the inside of the skull, too….and quantitatively.
I’m such a dummy! Yes, I meant in the top 2%. I know this because of a brain scan. Apparently most or all of that volume is dedicated to something other than math ability. Just as there are some people who cannot read despite being otherwise reasonably intelligent, I cannot do simple math, nor can I even begin to comprehend music. Numbers just don’t compute with me, but geometry does, which may explain my pathfinding abilities.
I have read, however, that these data are compromised by preferential preservation of male skeletons (which have larger skull volumes).
This said, I wonder why skull volume of our own species has decreased since the Cro Magnon time.
I hesitate to question a Nobel laureate, but how can the authors be sure that the observed distributions of these genetic variants do not reflect merely the idiosyncratic sorting of ancestral polymorphisms retained across closely spaced nodes in the organismal tree? There is even a term for this phenomenon: “hemiplasy”, formally defined as the topological discordance between a gene tree and a species tree attributable to lineage sorting of genetic polymorphisms that were retained across successive nodes in a species (or population) tree. If hemiplasy is involved, at least in part, then perhaps it might not be absolutely necessary to invoke post-separation hybridization and introgression to account for the observed distributions of alleles in these three human-like lineages. Just a thought.
For interested readers, more on hemiplasy can be found in my 2008 paper entitled “Hemiplasy: a new term in the lexicon of phylogenetics.”
Thank you! I will try to read your paper.
Very interesting! The Denisovian-to-modern-human transfers seem likely to be adaptive, given the connection to cold climates and high-altitudes. The Neanderthal-to-modern-human transfers are more of a stretch. Let’s suppose that those gene transfers are also adaptive. How long might it take for them to become fixed in the modern population? That would, minimally, depend on their contribution to fitness, but it could also depend on many other factors: the vagaries of population sizes over the millennia, losses in some populations that may have become extinct, how changing environments or migrations affect fitness of those genes, etc. Would fixation of favorable variants even be expected in these cases? Seems like air would be difficult to rule the Neanderthal genes either in *or* out as being adaptive in modern humans.
Incidentally, your point about back-crossings being fertile and viable makes a strong case for considering all three populations as belonging to the same species.
Thank you for the excellent science post!
As a quick answer to John Avise’s question about incomplete lineage sorting, there are indeed many old shared ancestral polymorphisms found in Neandertals or Denisovans and some living people; ABO blood types are an example. Introgressed haplotypes are different from shared ancestral polymorphisms in a few ways. The most relevant of these is haplotype length: polymorphisms that have been retained in two lineages for more than 600,000 years have as much as 10 times more recombination in their gene trees when compared to haplotypes that introgressed around 60,000 years ago. That means that introgressed haplotypes are usually a lot longer than the shared haplotypes that may flank ancestral polymorphisms. However these distributions do overlap (since Neandertals and Denisovans themselves had genetic variation) and statistical tests cannot always distinguish any particular shared haplotype as introgressed versus incomplete lineage sorting. The method of comparison with different African groups as discussed by Phil Ward above provides a complementary observation that helps to distinguish introgressed from ILS haplotypes. In papers like the one discussed here, authors commonly have applied a combination of statistics and focus on those with the highest confidence of introgression.
Thank you, John, for this response, which is very helpful and enlightening.
Coincidentally, my wife yesterday completed making a fun glass mosaic using a graphic representation of DNA as her inspiration. “Each twist of DNA also includes a transistor that suggests an intentional mutation.”
https://x.com/Jon_Alexandr/status/1762965677961511239
My wife was at first tickled to learn from 23andMe about her genetic heritage, which seemed to include more Neanderthal genes than the average person. She was somewhat crestfallen when 23andMe later re-calculated the amount to suggest a more average Neanderthal contribution. (Her background is primarily European.) In any case, she loves to participate in studies that correlate some of her quirks — like a strong dislike of cilantro — to her DNA record.