by Greg Mayer
UPDATE. A couple of readers have drawn attention to the website, gcbias, of Graham Coop, a population geneticist at UC Davis. He has excellent discussions, with nice graphics, of issues in genetic genealogy, including calculation of the number of “genetic units” in particular generations. As an example, 7 generations back you have 256 ancestors, but only 286 genetic units produced by recombination, so although, on average, you will have a chunk from each of those 256, it is entirely plausible to have zero (since inheritance is stochastic). It’s well worth browsing, and this and this are good places to start. (Thanks to rich lawler and S. Joshua Swamidass for the pointers.)
In February, I posted the syllabus for a seminar class entitled “Human Phylogeography” that I was teaching with my colleague Dave Rogers. The seminar was based primarily on a close reading of David Reich’s (2018) Who We Are and How We Got Here (published by OUP in the UK). Well, the class has concluded now, and so I thought I’d report back on what happened.
First, I’d like to say that the class was a success. We had 16 students, double the most I’ve ever had in a number of similar seminar courses over the years, and the students were very successful in engaging with the subject in both written and oral contributions to the class. One of the students was a history major, and towards the end of the semester a colleague in computer science mentioned that, quite coincidentally, he was reading the book, so he joined the class for the last few meetings. In many ways, it was what college is supposed to be like (though too often isn’t). I hope the students learned a lot. I did, and here is the first of the three most striking things I learned.
1. Recombination is a lot rarer than you think.
If you think back to the last time you studied genetics, you’ll recall the phenomenon of recombination, one aspect of which is crossing over. Crossing over occurs during meiosis. Chromosomes come in homologous pairs (23 pairs in humans, for 46 total), and in meiosis the homologues can exchange pieces with one another. The chromosomes physically touch and cross one another, which is observable under the microscope, and are called, appropriately enough, chiasmata (chiasma, sing.)
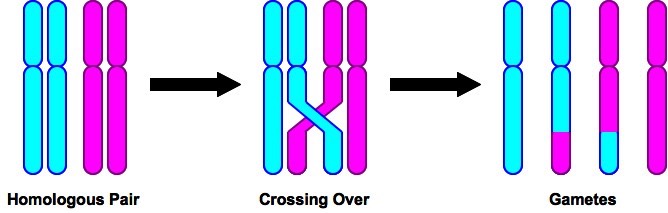
Recombination is important for a variety of reasons (for one, it increases genetic variability), but for our current purposes its importance is that it breaks up the nuclear genome from 23 genetic units into more, and smaller, units (as opposed to the mitochondrial genome, which has a number of genes, but all are inherited as a single genetic unit, since there is no recombination in mitochondria). In humans, it turns out, there are only 1-2 crossovers per chromosome per generation (1.2 per chromosome in fathers, 1.8 in mothers).
Now, I’d always thought that crossing over occurred frequently enough that we could think of the genome as essentially infinitely divisible. (There are 3 billion base pairs in the human genome, so, in the limit, there would be 3 billion genetic units, so not quite infinite!) But, it turns out that crossovers occur sufficiently infrequently that there is an appreciable chance that, if you go enough generations back, you share NO genes with your ancestor. This is because the number of ancestors goes up fast (2, 4, 8, 16, 32, 64, 128, 256, etc.), but the breaking up of the genome into smaller units by crossing over isn’t fast enough to ensure that the probability of sharing nothing is near zero.
Here’s a figure from Reich’s book showing how blocks of genes are broken up by recombination.

You start with an entirely Neanderthal chromosome (dark), which enters the anatomically modern human population by hybridization. A few generations later, the Neanderthal chromosome has been broken up, but it still occurs as largish blocks amongst the anatomically modern sections (gray). Still later, the blocks are smaller and fewer. (We’re assuming continued backcrossing into the anatomically modern population, so the % Neanderthal decreases; there could also be selection causing changes in the frequency of Neanderthal alleles). Finally, a present day individual has his Neanderthal DNA broken up into even smaller bits.
Here’s a figure from a talk by Svante Pääbo, showing in the top row for each chromosome (there are 22 listed, from 1-22) the entire genome of “Oase Boy” from 40K years ago in Romania. The green lines are Neanderthal sites in his genome. The five rows below Oase Boy are five modern human individuals; the colored lines are their “Neanderthal bits”. Note that for each chromosome, Oase boy has the biggest block of Neanderthal genes (green fluorescece):

Because of the age of the Oase sample, some of the black lines are missing data, and so Pääbo infers that there are seven large continuous blocks of Neanderthal genes (yellow bars above the Oase Boy line). Note that the modern individuals have less Neanderthal DNA, and it is not in large blocks.
Because the size of the blocks breaks up in a statistically predictable fashion, you can get a “recombination clock“, so that based on the size of the blocks you can estimate how many generations ago the hybridization occurred. For Oase Boy, Pääbo estimated that his Neanderthal ancestor occurred 4-6 generations back (his great great, or great great great, or great great great great grandfather).

Because the placement and frequency of crossing over is stochastic (random), the situation must be statistically modeled to derive sound estimates, and there will be a range of plausible estimates. And, since some of the fossils are well dated by other means, we can also estimate the long term human generation time, as was done by Priya Moorjani and her colleagues: it’s 26-30 years.
So, the low rate of recombination allows us to construct a “recombination clock”, and to estimate generation times. This is great stuff!
It also solved for me what was a puzzle. You may recall that last year Elizabeth Warren released the results of DNA tests showing that she had American Indian ancestry several generations back. This essentially confirmed what her family’s oral history said. The amount of her Indian ancestry was small (less than 1%), and a range of generations (6-10) was provided by the analysis (as was done by Pääbo for Oase Boy).
Now, there are a number of ways which these ancestry tests can be criticized, one of the most difficult for them being that there are very few North American Indian genotypes in the database used, and thus “American Indian” relationship is indicated by relationship to Central and South American Indians. Some critics of Warren, however, made erroneous criticisms. She did not contend, as some accused her of, of saying the results showed she was Cherokee—with few if any Cherokee in the database, the ancestry tests could not determine this. (And tribal membership is a legal matter, anyway, not directly dependent on genetic similarity.)
But some critics said that the data were consistent with her having no Indian ancestry at all. I wondered how they could say that– there are 3 billion bp, and 1 % of that is still a very large number. But now I realize my error. There are very many fewer genetic units– more than 23, but a lot less than 3 billion!– due to low rates of recombination. And, because of this, if you go back several generations, there is an appreciable probability of sharing no DNA with an indubitable ancestor. I now believe the critics must have looked at the latter fact, and realized Warren may not have DNA from all of her ancestors, and thus suggested she may have no Indian ancestry. But their error is that in saying she may lack DNA from an ancestor, say, 8 generations back, they are invoking an a priori probability. But in Warren’s case, her DNA was examined, and showed that she did have Indian ancestry.
Gravel, S. 2012. Population genetic models of local ancestry. Genetics 191:607-619. pdf
Ho, S. Y., Chen, A. X., Lins, L. S., Duchêne, D. A., & Lo, N. 2016. The genome as an evolutionary timepiece. Genome Biology and Evolution 8: 3006–3010. pdf
Huff, C.D. et mult. 2011. Maximum-likelihood estimation of recent shared ancestry (ERSA). Genome Research 21:768-774. pdf
Moorjani P, Sankararaman S, Fu Q, Przeworski M, Patterson N, Reich D. 2016. A genetic method for dating ancient genomes provides a direct estimate of human generation interval in the last 45,000 years. Proceedings of the National Academy of Sciences USA 113:5652-7. pdf
Pääbo, Svante. 3 October 2018. A Neanderthal Perspective on Human Origins. (video: embedded below)
Reich, D. 2018. Who We Are and How We Got Here: Ancient DNA and the New Science of the Human Past. Pantheon, New York.

See you there.
Sub
Sub2
Fascinating read.
Very informative post, thanks. And your posts on education are always useful. Teachers are not taught how to facilitate learning in an interactive, constructive way that involves thinking and doing.
In myopic America, it’s ‘by the book’ or ‘following the rules’. Like Snoop says: ‘No inspiration, all day demonstration’. No wonder kids are turned off by education.
Sub
This sounds like the sort of seminar I would have enjoyed very much. Thanks for sharing!
Many thanks this was a very interesting read. I have always been interested in genetics, having haemophilia may be a reason for that. If you are going to be born with an awkward condition at least hope it is an interesting one.
I also hadn’t realised that crossovers were quite that rare. So in other words, in each chromosome pair, one chromosome is likely to be mostly from your paternal grandfather or mostly from your paternal grandmother, while the other is similar on the maternal side. Hence each chromosome you send to a child can come mostly from just one grandparent.
Is the behaviour of each chromosome pair in meiosis independent of each other pair? Each grandparent gives you around 6 chromosomes, but is this completely random, i.e. multinomial(23,1/4,1/4,1/4,1/4)? Or does meiosis “cluster” so that most chromosomes can come from the same grandparent?
In the first pic in the OP, you’ll see that when there is a cross over, it involves two chromatids, while two chromatids are unrecombined. So, if there’s a single crossover, the resulting 4 gametes will be 1 with the paternal, 1 with the maternal, and 2 with(complementarily) recombined chromosomes. There can be multiple cross overs, which could involve the same two chromatids twice, or both maternal with both paternal, etc. So, with 1 crossover you’re sure to get 1 paternal and 1 maternal gamete; with multiple crossovers you might get them, but as the number of cross overs goes up, the chances go down.
Each homologous pair assorts independently, though– this is one of Mendel’s ‘laws’. So, there’s no tendency for paternal chromosomes to wind up in the same gamete. For humans, with 22 autosomal chromosome pairs, and exactly one crossover per pair, the probability of getting all paternal chromosomes in a gamete is .25^22, and there would be the same probability of getting all maternal. (One of the 4 chromatids in each homologous chromosome pair is paternal, so the probability is 1/4 or .25 of a gamete getting it; now repeat this for all 22 chromosome pairs.) That’s a very small probability.
So, meiosis does not cluster chromosomes from the same grandparent.
GCM
There are some interesting implications of there being only 1 – 2 recombinations per chromosome + recombination occurring at the 4-strand stage (to me and, therefore, to my genetics students, anyway).
Even though most of your chromosomes are a mixture of the two grandparents’ on that side (mom’s side or dad’s side), it is likely that at least a few of your chromosomes are an intact copy identical to a chromosome in one of your grandparents. More of these should be on dad’s side, because of the higher recombination rate in females. This happens when a chromatid not involved in any recombination gets packaged into the gamete that became you.
Also, you are *certain* to share 1/2 of your DNA with mom and 1/2 with dad (not counting XY), but you only *on average* share 1/4 of your DNA with any particular grandparent.
Some of the best discussions on the probability of sharing chromosomal chunks with your maternal and paternal lineages are posted on Graham Coop’s blog, gcbias. He has a whole series on genomics and genealogy and it is excellent!
See here, for example:
https://gcbias.org/2013/10/20/how-much-of-your-genome-do-you-inherit-from-a-particular-grandparent/
and here:
https://gcbias.org/2013/11/04/how-much-of-your-genome-do-you-inherit-from-a-particular-ancestor/
Good question
Good question
Sub
Very interesting read.
I always assumed that during recombination the DNA is much more likely to “break” between genes that inside them, because breaking up a gene is risky and probably punished by natural selection. Therefore I always assumed that for the purpose of recombination the genome is divisible to 30 000 + some parts. Non-coding or junk parts could be easier to break, but still, I thought it is in the magnitude of 100 000 or so top, with genes virtually never breaking inside.
From the description above it is not clear for me whether I was totally wrong on the matter.
Other than this, I also overestimated the frequency of recombination and that is actually weird from me. I knew that linkage disequilibrium can remain in mixed populations even after millennia, but never actually though through what recombination frequency is implied by this fact.
There certainly is intragenic recombination. It’s rarer than intergenic because i) every intragenic recombination affects one gene, but is intergenic for many genes either side of the crossover; and ii) much of the DNA has no genes in it, and so crossovers in that DNA can’t be intragenic. Recombination rates vary widely across spots on the chromosomes; there are “recombinational hotspots” with much higher rates (though still in less than 1% of meioses). I don’t know if this variation in rate includes any tendency for cross overs not to occur within genes.
GCM
That’s an interesting point. You would expect that it would be random, ignoring gene boundaries, on the principle that mutations (and recombinations?) are random but natural selection isn’t.
However, if there is genetic influence on recombination rates and/or hot-spots then this might not be true. Recombination in the middle of a critical gene might be selected against.
Maybe this will be discussed in later posts?
This was most informative; thanks for posting it. I am looking forward to the next installments. However, I am not sure of your conclusion “rarer than you think”. A Google images search for “crossing over” shows that the majority of diagrams resemble the one in the post with just one chiasma per 4 chromatids, if anything an underestimation of the extent of recombination.
General biology textbooks almost always show a single chiasma because they are trying to diagram it simply for students to understand. I never thought myself that this might be an indication of frequency, but it turns out it’s not far off for humans.
Genetics textbooks usually include a discussion of how to do genetic mapping through recombination studies, and double and other multiple crossovers must be looked at to do this, and there are usually accompanying diagrams depicting the multiple crossovers.
Here’s an homologous pair with 5 chiasmata (crossovers; from https://web.archive.org/web/20160728190019/http://users.rcn.com/jkimball.ma.ultranet/BiologyPages/M/Meiosis.html)
https://web.archive.org/web/20160622021259im_/http://users.rcn.com/jkimball.ma.ultranet/BiologyPages/C/Chiasmata013.jpg
GCM
Did she really have Indian DNA or was it just a Mesoamerican proxy for Indian DNA?
In the study of her DNA, the evidence is quite strong for what is termed “Native American”. Because there are relatively few United States American Indians in the database, I would assume that her 5 Native American segments identified in the study are widely shared in the Americas, but I don’t know the details of the phylogeography for these segments. It is conceivable that she has a Meso-American Indian ancestor, but this seems less likely to me.
GCM
There is certainly a good chance that someone from Oklahoma could have some American Indian DNA. Intermarriage was not uncommon especially with the Cherokee. (Not saying she has Cherokee DNA, it is just an example.) John Ross, the chief of the Cherokee Nation at the time of removal to Oklahoma was only 1/16th Cherokee.
In other states, like Missouri and Kentucky, however, most claims of Indian, and especially Cherokee ancestry are based on myths about what happened during the Trail of Tears.
Mesoamericans are Indians.
No, it does not. Warren’s family lore makes specific claims:
a) a single Cherokee ancestor, a great-great-great-grandmother named O.C. Sarah Smith;
b) Her mother was Cherokee on one side and Delaware on the other (thus, two Indian ancestors);
c) Her parents eloped in 1932 because her racist paternal grandparents refused to let her father marry her mother, who was known to be “part Cherokee and part Delaware”.
Cherokee Genealogist Twila Barnes has exhaustively researched Warren’s ancestry and shown all these claims to be false. Not a single Warren ancestor stretching back 5 generations is listed as Indian in any census or on any official document.
http://www.pollysgranddaughter.com/p/elizabeth-warren-information.html
As the OP notes, Warren’s trace genetics cannot be identified by tribe (or even as North American, for that matter.) Based on the figures provided in the OP, the ancestor Warren inherited her <1% New World genes from was born c. 156 – 300 years before her, between c. AD 1649 – 1793.
The mysterious O.C. Smith (no “Sarah”) gave birth to Warren’s G-G-grandfather, Preston H. Crawford, in 1824. No records list O.C. Smith-Crawford as Indian.
The Trail of Tears, the forced relocation of the Cherokee to Oklahoma, took place in 1838-9.
Interestingly, Warren’s G-G-G-grandfather, William J. Crawford, helped round up Cherokees for the Trail of Tears. He and his wife remained in Tennessee.
***
Based on her spurious family lore, Warren also claimed Federal minority status on official documents. As she has no ancestors listed on either the Dawes Roll (Cherokee Nation, United Keetoowah Band of Cherokee) first compiled in 1893, or the Baker Roll of 1924 (Eastern Band of Cherokee Indians), Warren is not eligible for Cherokee citizenship, thus was could not legally claim minority status.
I saw an article on Carl Zimmer’s book “She Has Her Mother’s Laugh: The Powers, Perversions, and Potential of Heredity.”
One thing noted from the book was that the chance of you having any DNA from an ancestor 10 generations back is about 50%. The reason given is just as you have described here.
There is something very strange about having an ancestor and descendant 10 generations apart that are related in the usual sense of the word, but share no DNA. Are they really related at that point?
Are they really related at that point? I’d say yes, but it would be difficult to prove.
Presumably the 50% figure refers to the odds of directly inheriting DNA from that 10th-generation ancestor. But failure to do so doesn’t mean you share no DNA; it just means your shared DNA came to you both from an ancestor even farther back.
I inherited zero DNA from my cousins, but that doesn’t mean we’re not related. And we all share 98% of our DNA with chimps.
Yes, cousin Gregory, you are right, we are all related if you look back far enough. And we share plenty of DNA sequences anyway; we can’t be that different from each other.
It still seems odd to say that someone is a direct descendant of an ancestor 10 generations ago, when they didn’t get any of their DNA from that ancestor. It’s not really a blood line if there is nothing that has been passed along.
Do we have any understanding of how this stochasticity is achieved? Is it possible that quantum effects play a role here?
With some trepidation (considering the context here), genetic ghosts are a key plank in the argument in the case I made on the Adam and Eve. Curious your thoughts. https://asa3.org/ASA/PSCF/2018/PSCF3-18Swamidass.pdf
Figure 1 might be helpful.
Graham Coop’s series on this is quite good too. https://gcbias.org/2017/12/19/1628/
Thanks for sharing this Greg – it is still in my ‘to read’ piles! (plural!)
I too thought that crossing over happened a lot, since discussing it was a big deal in General Biology I and related materials. Guess not. 🙂